Whole genome sequencing (WGS) in food microbiology is the process of determining the whole genome sequence of a single cultured isolate (e.g. a bacterial colony, a virus or any other organism) at a single time. WGS rely on next-generation sequencing (NGS) which made it possible to massively parallel sequence small, overlapping stretches of DNA in one reaction. Sequencing can be done through short read sequencers (50-500bp) with low error rate (high accuracy) that will lead to incomplete draft genomes which can be used for comparative genomics and phylogeny. Sequencing can also be done by long read sequencing (1k to 100kb) with a high error rate, but which can lead to complete genome. These millions of DNA reads are then assembled to reconstitute the whole genome and analyzed to compare it to known sequences through bioinformatics. When applied to a biological sample (e.g. a microbial population instead of an isolate), it is called metagenomics and metabarcoding.
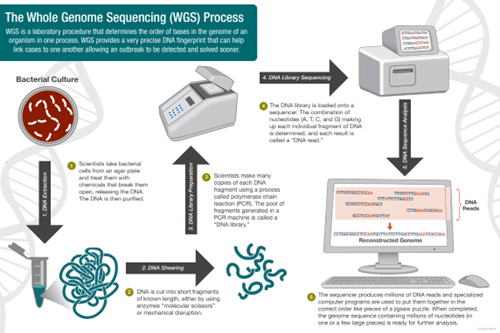
Figure 1: WGS workflow (CDC)
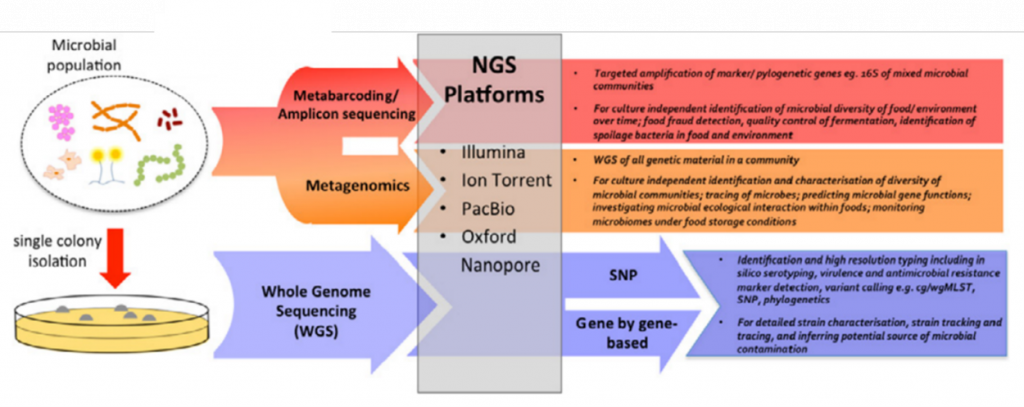
Figure 2: Jagadeesan et al.
Regarding the bioinformatic analysis, there are two complementary bioinformatic approaches for comparative genomics:
| DNA base approach Single nucleotide polymorphism (SNP) |
Gene-by-gene approach Multi-locus sequence typing either a defined core genome (cgMLST) or the whole genome (wgMLST) |
|---|---|
| Comparison to a carefully selected reference genome from a closely related strain | Comparison to a reference database of all known gene variants (alleles), from numerous strains for a particular species. Public validated databases available for: Listeria monocytogenes, Salmonella, Escherichia/Shigella, Yersinia, Campylobacter |
| Difficult to standardize between labs. Adapted to centralized surveillance by one lab doing all the analyses | Easily standardized by using common databases. Adapted to decentralized surveillance in a network of labs that compare new isolates to a database. Adopted by PulseNet |
| Requires expert bioinformatic support to use open-source software. | Can be done with user-friendly commercial software |
| Confirm the relatedness between isolates in a cluster | Identify primary clusters |
Once the number of SNP/alleles differences has been determined, the results can be displayed as a phylogenetic tree to assess how closely related two isolates are genetically. As phylogeny reflects imperfectly epidemiological relatedness, it is crucial to use epidemiological evidence (patient interviews, traceability, regulatory inspection, evidence of breakdown of food safety measures,..) to support the phylogenetic findings, determine the food vehicle, the original source of contamination, and mode of transmission. The threshold to confirm that two isolates are closely related depends on the bacteria, the environment, the epidemiological context and the WGS analysis approach but typically 0–20 SNP/allele differences usually mean the two food pathogen isolates are highly related whereas 50–100 SNPs/alleles differences mean they are not likely coming from the same source.
A complementary article on WGS for food safety and on the Future of WGS is available here.
Source: The Use of Next Generation Sequencing for Improving Food Safety: Translation into practice
